Kane-Mele模型原始论文为:Z2 Topological Order and the Quantum Spin Hall Effect、Quantum Spin Hall Effect in Graphene。
这里考虑没有Rashba自旋轨道耦合的情况,哈密顿量为:
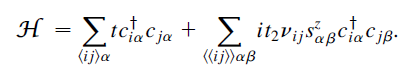
此时的哈密顿量可以分解成两个独立的Haldane模型。Haldane模型可以参考这篇:Haldane模型哈密顿量与能带图(附Python代码)。
自旋向上和自旋向下对应的Haldane模型给出相反的霍尔电导,整体满足时间反演对称性。
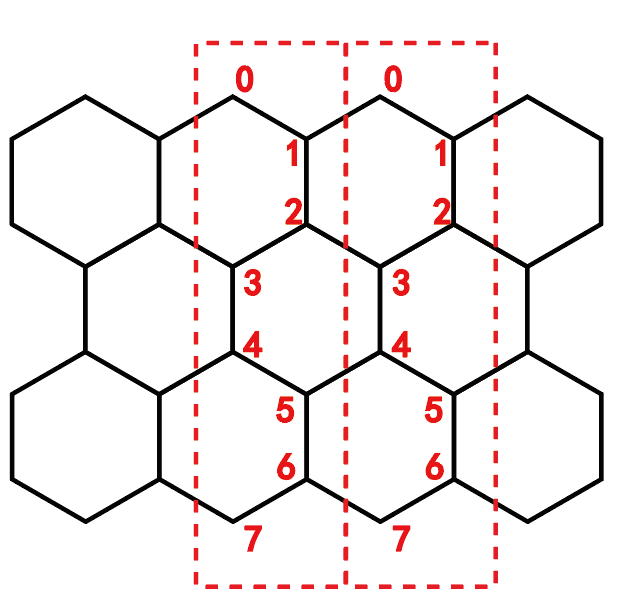
蜂窝格子的编号为:
准一维的Kane-Mele模型(不考虑Rashba自旋轨道耦合)能带图代码为:
"""
This code is supported by the website: https://www.guanjihuan.com
The newest version of this code is on the web page: https://www.guanjihuan.com/archives/4829
"""
import numpy as np
import matplotlib.pyplot as plt
from math import *
import cmath
import functools
def hamiltonian(k, N, M, t1, t2, phi): # Kane-Mele model
# 初始化为零矩阵
h00 = np.zeros((2*4*N, 2*4*N), dtype=complex) # 因为自旋有上有下,所以整个维度要乘2。这里4是元胞内部重复单位的大小,规定元胞大小以4来倍增。
h01 = np.zeros((2*4*N, 2*4*N), dtype=complex)
for spin in range(2):
# 原胞内的跃迁h00
for i in range(N):
# 最近邻
h00[i*4*2+0*2+spin, i*4*2+1*2+spin] = t1
h00[i*4*2+1*2+spin, i*4*2+0*2+spin] = t1
h00[i*4*2+1*2+spin, i*4*2+2*2+spin] = t1
h00[i*4*2+2*2+spin, i*4*2+1*2+spin] = t1
h00[i*4*2+2*2+spin, i*4*2+3*2+spin] = t1
h00[i*4*2+3*2+spin, i*4*2+2*2+spin] = t1
# 次近邻
h00[i*4*2+0*2+spin, i*4*2+2*2+spin] = t2*cmath.exp(-1j*phi)*sign_spin(spin)
h00[i*4*2+2*2+spin, i*4*2+0*2+spin] = h00[i*4*2+0*2+spin, i*4*2+2*2+spin].conj()
h00[i*4*2+1*2+spin, i*4*2+3*2+spin] = t2*cmath.exp(-1j*phi)*sign_spin(spin)
h00[i*4*2+3*2+spin, i*4*2+1*2+spin] = h00[i*4*2+1*2+spin, i*4*2+3*2+spin].conj()
for i in range(N-1):
# 最近邻
h00[i*4*2+3*2+spin, (i+1)*4*2+0*2+spin] = t1
h00[(i+1)*4*2+0*2+spin, i*4*2+3*2+spin] = t1
# 次近邻
h00[i*4*2+2*2+spin, (i+1)*4*2+0*2+spin] = t2*cmath.exp(1j*phi)*sign_spin(spin)
h00[(i+1)*4*2+0*2+spin, i*4*2+2*2+spin] = h00[i*4*2+2*2+spin, (i+1)*4*2+0*2+spin].conj()
h00[i*4*2+3*2+spin, (i+1)*4*2+1*2+spin] = t2*cmath.exp(1j*phi)*sign_spin(spin)
h00[(i+1)*4*2+1*2+spin, i*4*2+3*2+spin] = h00[i*4*2+3*2+spin, (i+1)*4*2+1*2+spin].conj()
# 原胞间的跃迁h01
for i in range(N):
# 最近邻
h01[i*4*2+1*2+spin, i*4*2+0*2+spin] = t1
h01[i*4*2+2*2+spin, i*4*2+3*2+spin] = t1
# 次近邻
h01[i*4*2+0*2+spin, i*4*2+0*2+spin] = t2*cmath.exp(1j*phi)*sign_spin(spin)
h01[i*4*2+1*2+spin, i*4*2+1*2+spin] = t2*cmath.exp(-1j*phi)*sign_spin(spin)
h01[i*4*2+2*2+spin, i*4*2+2*2+spin] = t2*cmath.exp(1j*phi)*sign_spin(spin)
h01[i*4*2+3*2+spin, i*4*2+3*2+spin] = t2*cmath.exp(-1j*phi)*sign_spin(spin)
h01[i*4*2+1*2+spin, i*4*2+3*2+spin] = t2*cmath.exp(1j*phi)*sign_spin(spin)
h01[i*4*2+2*2+spin, i*4*2+0*2+spin] = t2*cmath.exp(-1j*phi)*sign_spin(spin)
if i != 0:
h01[i*4*2+1*2+spin, (i-1)*4*2+3*2+spin] = t2*cmath.exp(1j*phi)*sign_spin(spin)
for i in range(N-1):
h01[i*4*2+2*2+spin, (i+1)*4*2+0*2+spin] = t2*cmath.exp(-1j*phi)*sign_spin(spin)
matrix = h00 + h01*cmath.exp(1j*k) + h01.transpose().conj()*cmath.exp(-1j*k)
return matrix
def sign_spin(spin):
if spin==0:
sign=1
else:
sign=-1
return sign
def main():
hamiltonian0 = functools.partial(hamiltonian, N=20, M=0, t1=1, t2=0.03, phi=pi/2)
k = np.linspace(0, 2*pi, 300)
plot_bands_one_dimension(k, hamiltonian0)
def plot_bands_one_dimension(k, hamiltonian):
dim = hamiltonian(0).shape[0]
dim_k = k.shape[0]
eigenvalue_k = np.zeros((dim_k, dim))
i0 = 0
for k0 in k:
matrix0 = hamiltonian(k0)
eigenvalue, eigenvector = np.linalg.eig(matrix0)
eigenvalue_k[i0, :] = np.sort(np.real(eigenvalue[:]))
i0 += 1
for dim0 in range(dim):
plt.plot(k, eigenvalue_k[:, dim0], '-k')
plt.ylim(-1, 1)
plt.show()
if __name__ == '__main__':
main()
计算出的能带图为:
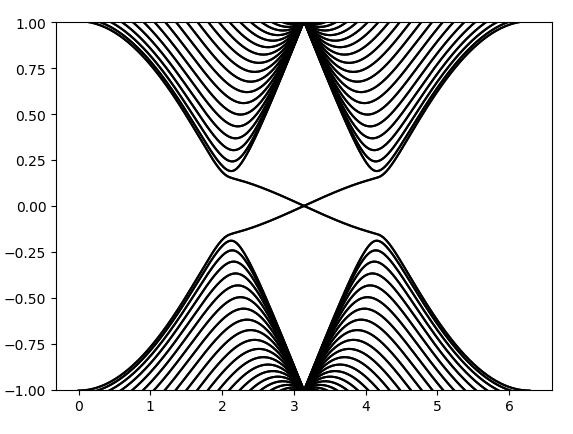
这里画出来的能带和Haldane模型对应参数的能带看起来完全一样,但这里的能带是二重简并的,且有不同的拓扑性质。这里的拓扑性质由Z2不变量来定义。
关于详细的讨论以及考虑Rashba自旋轨道耦合的情况,可以看原始论文或者相关综述。
实验上首先发现该量子自旋霍尔效应的是在Mercury Telluride Quantum Wells中,该体系由BHZ模型来描述,参考这篇:BHZ模型哈密顿量与能带图(附Python代码)。
附Matlab代码(由Zhong-Fu Li同学提供):
% Purpose: Plot Kane-Mele's bands
% Date: Dec 07, 2021 @HNU
% Author: Ji-Huan Guan;
% Modify by Zhong-Fu Li;
% More code can be found in https://www.guanjihuan.com/
close all
clc
clear all
tic;
k0 = linspace(0,2*pi,100);
for kk = 1:100
k = k0(kk);
H1 = HH(k);
[VV,DD] = eig(H1);
Ds(:,kk) = sort(diag(DD),'ascend'); % sort eigenvalue
end
plot(k0/2/pi,Ds,'k.');
ylim([-1 1])
set(gca,'xtick',[0:0.5:1])
set(gca,'XTickLabel',{'\bf 0','\bf \pi','\bf 2\pi'})
set(gcf,'color','w');
set(gca,'fontsize',16,'LineWidth',1.1);
ylabel('Energy(a.u.)','fontname','Arial');
toc
% define Hamiltonian
function H = HH(k)
N = 10;
M = 0;
t1 = 1;
t2 = 0.03;
phi = pi/2;
h00 = zeros(2 * 4 * N, 2 * 4 * N);
h01 = zeros(2 * 4 * N, 2 * 4 * N);
for spin = 1:2
for ii = 0:N-1
% nearest neighbor couplings
h00(ii * 4 * 2 + 0 * 2 + spin, ii * 4 * 2 + 1 * 2 + spin) = t1;
h00(ii * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 0 * 2 + spin) = t1;
h00(ii * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 2 * 2 + spin) = t1;
h00(ii * 4 * 2 + 2 * 2 + spin, ii * 4 * 2 + 1 * 2 + spin) = t1;
h00(ii * 4 * 2 + 2 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = t1;
h00(ii * 4 * 2 + 3 * 2 + spin, ii * 4 * 2 + 2 * 2 + spin) = t1;
%next nearest neighbor couplings
h00(ii * 4 * 2 + 0 * 2 + spin, ii * 4 * 2 + 2 * 2 + spin) = t2 * exp(-1j * phi) * sign_spin(spin);
h00(ii * 4 * 2 + 2 * 2 + spin, ii * 4 * 2 + 0 * 2 + spin) = conj(h00(...
ii * 4 * 2 + 0 * 2 + spin, ii * 4 * 2 + 2 * 2 + spin));
h00(ii * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = t2 * exp(-1j * phi) * sign_spin(spin);
h00(ii * 4 * 2 + 3 * 2 + spin, ii * 4 * 2 + 1 * 2 + spin) = conj(h00(...
ii * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin));
%
end
for ii = 0:N-2
% nearest neighbor couplings
h00(ii * 4 * 2 + 3 * 2 + spin, (ii + 1) * 4 * 2 + 0 * 2 + spin) = t1;
h00((ii + 1) * 4 * 2 + 0 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = t1;
% next nearest neighbor couplings
h00(ii * 4 * 2 + 2 * 2 + spin, (ii + 1) * 4 * 2 + 0 * 2 + spin) = t2 *exp(1j * phi) * sign_spin(spin);
h00((ii + 1) * 4 * 2 + 0 * 2 + spin, ii * 4 * 2 + 2 * 2 + spin) = conj(h00(...
ii * 4 * 2 + 2 * 2 + spin, (ii + 1) * 4 * 2 + 0 * 2 + spin));
h00(ii * 4 * 2 + 3 * 2 + spin, (ii + 1) * 4 * 2 + 1 * 2 + spin) = t2 * exp(1j * phi) * sign_spin(spin);
h00((ii + 1) * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = conj(h00(...
ii * 4 * 2 + 3 * 2 + spin, (ii + 1) * 4 * 2 + 1 * 2 + spin) );
end
% hopping of intercell h01
for ii = 0:N-1
% nearest neighbor couplings
h01(ii * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 0 * 2 + spin) = t1;
h01(ii * 4 * 2 + 2 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = t1;
% next nearest neighbor couplings
h01(ii * 4 * 2 + 0 * 2 + spin, ii * 4 * 2 + 0 * 2 + spin) = t2 * exp(1j * phi) * sign_spin(spin);
h01(ii * 4 * 2 + 1 * 2 + spin, ii* 4 * 2 + 1 * 2 + spin) = t2 * exp(-1j * phi) * sign_spin(spin);
h01(ii * 4 * 2 + 2 * 2 + spin, ii * 4 * 2 + 2 * 2 + spin) = t2 * exp(1j * phi) * sign_spin(spin);
h01(ii * 4 * 2 + 3 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = t2 * exp(-1j * phi) * sign_spin(spin);
h01(ii * 4 * 2 + 1 * 2 + spin, ii * 4 * 2 + 3 * 2 + spin) = t2 * exp(1j * phi) * sign_spin(spin);
h01(ii * 4 * 2 + 2 * 2 + spin, ii * 4 * 2 + 0 * 2 + spin) = t2 * exp(-1j * phi) * sign_spin(spin);
if ii ~= 0
h01(ii * 4 * 2 + 1 * 2 + spin, (ii - 1) * 4 * 2 + 3 * 2 + spin) = t2 * exp(1j * phi) * sign_spin(spin);
end
end
for ii = 0:N-2
h01(ii * 4 * 2 + 2 * 2 + spin, (ii + 1) * 4 * 2 + 0 * 2 + spin) = t2 *exp(-1j * phi) * sign_spin(spin);
end
end
H = h00 + h01 * exp(1j * k) + h01' * exp(-1j * k);
end
function sign = sign_spin(spin)
if spin == 1
sign = 1;
else
sign = -1;
end
end运行结果:
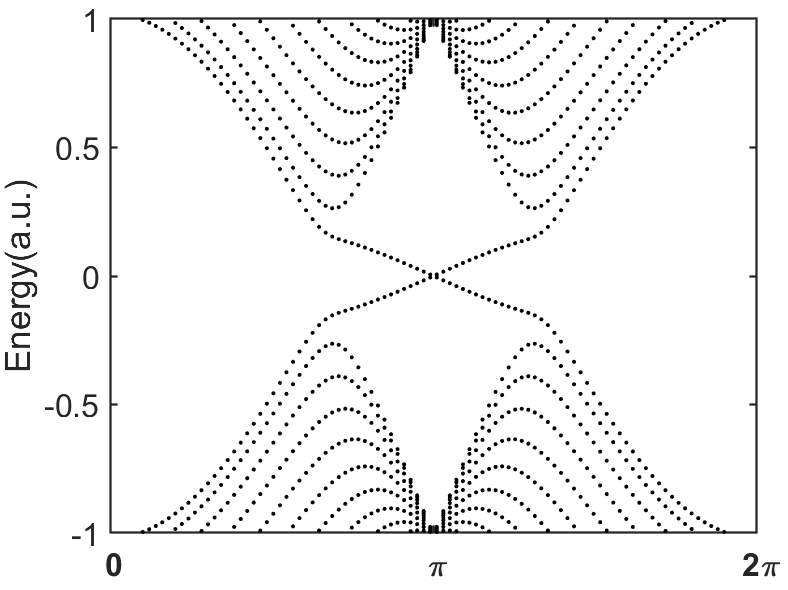
【说明:本站主要是个人的一些笔记和代码分享,内容可能会不定期修改。为了使全网显示的始终是最新版本,这里的文章未经同意请勿转载。引用请注明出处:https://www.guanjihuan.com】
这里的参数M指什么?
质量项,或交错势。这里的代码没有用到,忘了删除了,暂且保留,有需要考虑这一项时可以在代码中补充。
你好,请问MATLAB代码中最后的函数
function sign = sign_spin(spin)
if spin == 0 这行是不是应该写成 if spin == 1,其他不变
sign = 1;
else
sign = -1;
end
是的。感谢提醒,已更正。
真的很感谢老师分享的这些文章,从中学到了很多非常有用且能让我醍醐灌顶的知识。让我能在短时间内把Kane-Mele模型的锯齿型和扶手型边界的能带图重复出来!
你好,可以分享一下你重复的锯齿形和扶手椅形边界的能带图吗
有没有用matlab画这个图的代码,没有学过Python
没有的。主要意思其实还是能大概看出来,可以自己改写成Matlab
已更新matlab代码。
想问一下Rashba效应项怎么写,看论文没看明白?
其实我也还没怎么看,没做这方面的研究。不懂就多看几遍吧,或者看看其他文献中的表述。
谢谢。我想再问您一个问题,在对实空间哈密顿量作傅里叶变化时,写成h00 + h01*cmath.exp(1j*k) +h01.transpose().conj()*cmath.exp(-1j*k)道理何在呢?我不太明白这样做整体傅里叶变换。
这个形式是傅里叶变换后的的表达式,中间推导过程略掉了。这样取元胞后,整体类似于一维的方格子链。傅里叶变换参考这篇:离散格子的傅里叶变换
好的,十分感谢